Welcome to MeSiteIG
About MeSiteIG
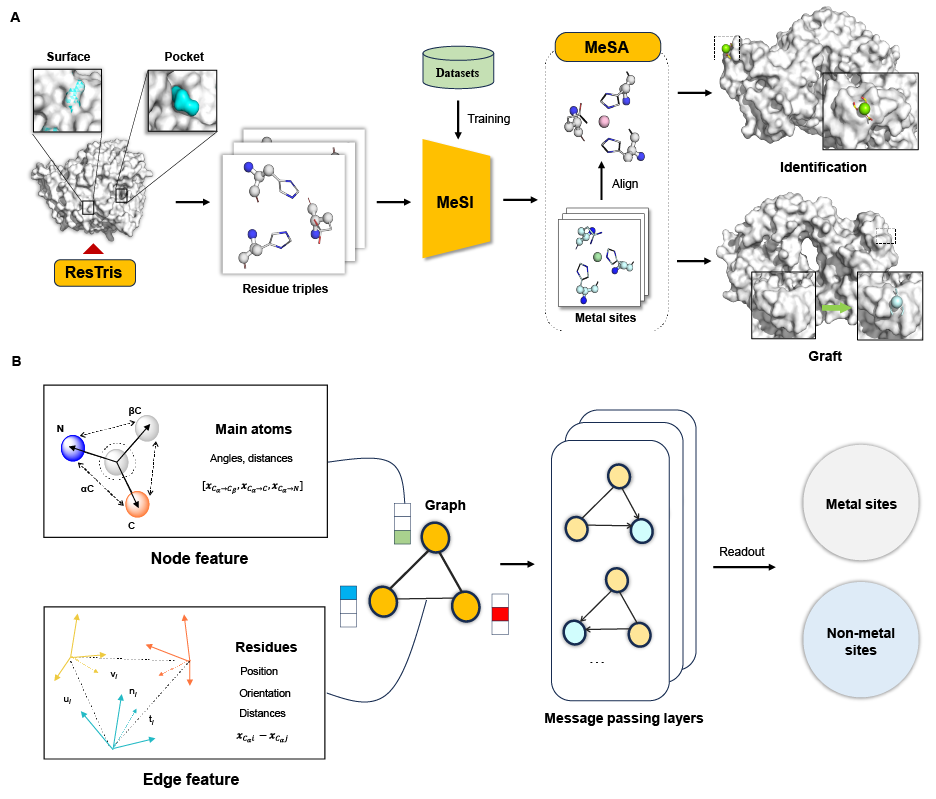
MeSiteIG is a versatile, residue-triplet-based tool for identifying metal sites. It integrates graph neural networks (GNNs) and physics-based matching algorithms to achieve fast and accurate identification. It represents each residue as a node in a graph, employing a three-node graph, characterized by scalar and vector features of nodes and edges, to depict a residue triplet. MeSiteIG provides a framework for predicting potential and graftable metal sites in protein PDB files, offering grafting solutions for the latter.
MeSiteIG is written in python and can be run in Linux and Windows. Details please see MeSiteIG Manual.
Please click to download MeSiteIG.zip file.
MeSiteIG is free for academic use, and please send user information (including PI name, institution, contact email, etc.) to ddtmlab_gbl@sina.com to obtain password to unzip the password-protected ZIP files.We sincerely are open to receiving support and advice from academic and industrial communities to improve MeSiteIG 's usefulness, please email us:ddtmlab_gbl@sina.com.
How to cite MeSiteIG
When using MeSiteIG, we kindly ask you to cite the article:
Jun-Lin Yu, Yao-Geng Wang, Jian Peng, Jing-Wei Wu, Cong Zhou, Guo-Bo Li*. Geometric deep learning-enabled metal site identification and grafting.
*To whom correspondence should be addressed.